|
DaGO-Fun - Database for GO-based Functional Annotation Analysis |
Browsing Tools
GO analysis Tools
- Measure Selection Guide
- Semantic Similarity
- Protein Identification
- Term Fuzzy-Enrichment
- Protein Classification
Functional Networks
Annotation Prediction
Browsing Resources
Protein Resources
Protein Interactions
Annotation Analysis
| GOSS-FEAT Description | |||||
Welcome to the description of the GOSS-FEAT tool allowing one to learn about the biology of an experiment explained by a list of genes or proteins by identifying functional annotation or Gene Ontology groups which are over-represented in this gene or protein list. GOSS-FEAT helps in the analysis of such lists and provides statistics about the GO terms contained in the list and ranks these GO annotations giving the most representative GO terms first. The tool provides a stepwise query selection menu, enabling the user to construct a query and adapting the selection choices in the process, leaving only relevant options open that correspond to his/her selections.
| |||||
| 1. User Input step and Result outputs | |||||
The user input is a list of UniProt protein accessions or gene names. In this case, input is UniProt protein accessions aligned and pasted in the Input text area or uploaded from a file. The user input is of the following form:
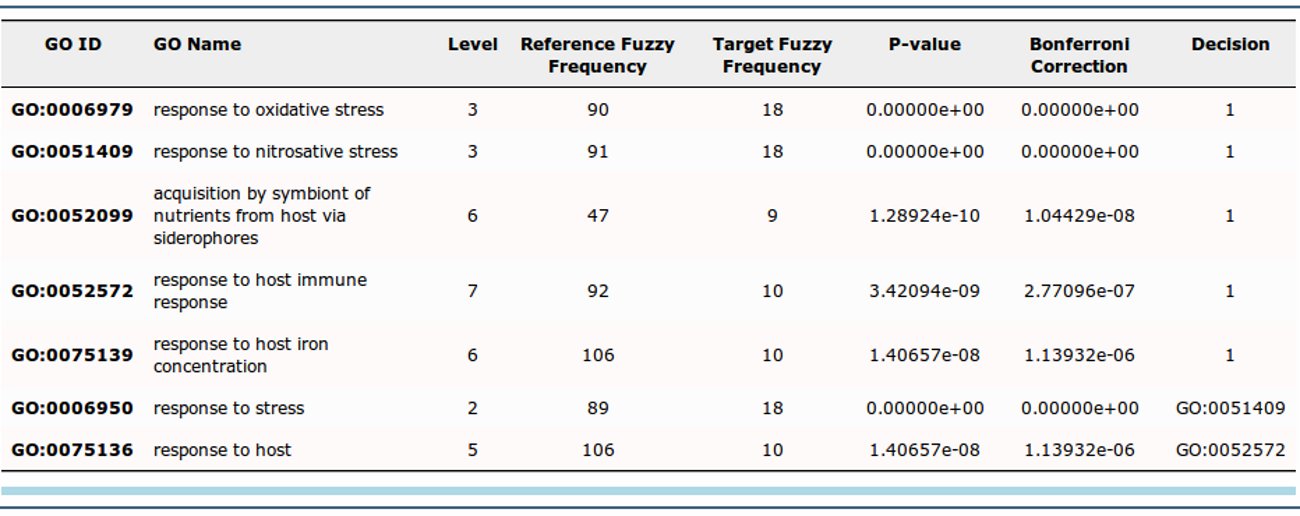
Depending on the term semantic similarity approach selected, the GOSS-FEAT tool produces a comprehensive summary in a table format on the next page of the user interface. The output list of GO terms is ranked by the p-value and can be limited by a p-value cutoff and this output is a table with eight columns as shown below:
Note that in all the DaGO-Fun applications, the level of a term is the length of the longest path (or the maximum number of links) from the root of the ontology down to that term. The root itself is located at the level 0 considered to be the reference level. Note that by clicking on a given active GO ID, the associated details about the term in the GO-DAG is displayed using the AmiGO tool. | |||||
| 2. Important Note: | |||||
A target list of at most 2000 protein UniProt accessions or gene names is recommended, in which case the over-represented terms identified are displayed with 10 per page. Unfortunately if you have cases where your data exceed these limitations, you can contact the administrators who are willing to collaborate and run large data sets for analysis. For more information, please refer to the associated publication: "Gaston K. Mazandu and Nicola J. Mulder. DaGO-Fun: Tool for Gene Ontology-based functional analysis using term information content measures, 2013", DaGO-Fun preliminary paper currently under review. |